Research
One common theme in our research is using networks to connect diverse data and provide a cohesive view of a biological process. We develop new computational methods and also work with collaborators to apply them to study specific conditions and diseases. As part of the John W. and Jeanne M. Rowe Center for Research in Virology at the Morgridge Institute, we are particularly interested in applications in viral infection and virus-induced cancers.
In addition, we create machine learning methods to guide biochemistry experiments, especially for protein engineering and drug discovery. Example studies include Gelman et al. 2021 and Alnammi et al. 2023.
Biological pathway reconstruction
 Figure from Köksal et al. 2018
Figure from Köksal et al. 2018
The timing of signaling events within cells can teach us how proteins pass messages to respond to changes in the cellular environment. Our Temporal Pathway Synthesizer (TPS) algorithm evaluates which possible signaling pathways could have generated an observed phosphorylation response. It uses constraints such as the timing of phosphorylation changes to rule out pathways in which late-responding proteins pass messages to (phosphorylate) early-responding proteins.
Protein engineering
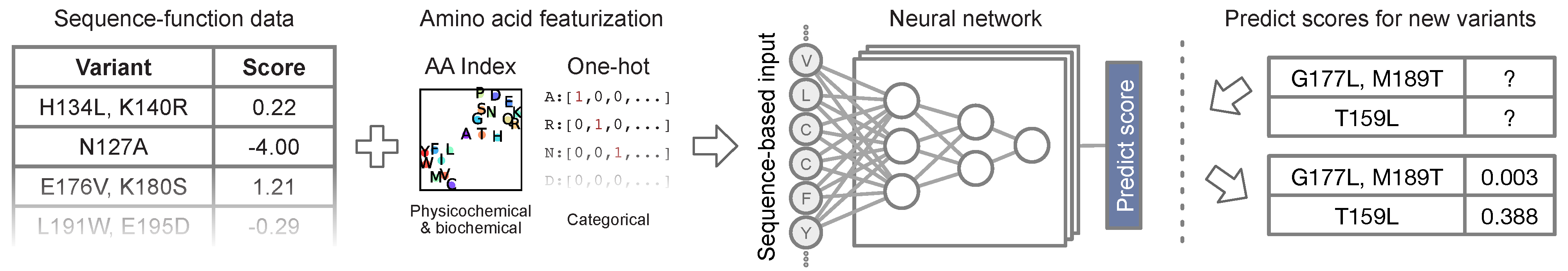 Figure from Gelman et al. 2021
Figure from Gelman et al. 2021
Protein engineering involves improving the function of a protein, such as its enzymatic activity. Our nn4dms software implements multiple neural network architectures for training models that predict quantitative protein function from protein sequence changes. METL improves on standard neural networks by combining biophysical modeling and protein language models. We collaborate with the Romero lab to design machine learning models and test protein designs in the wet lab.